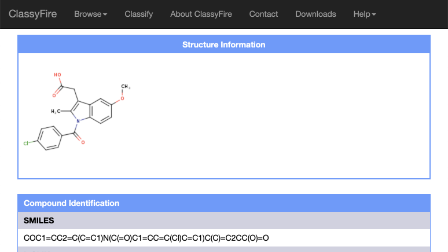
David Arndt
Bioinformatician and software developer with experience in the areas metabolomics, metagenomics, cheminformatics, and protein structural biology. Contributed to the development of several popular databases and tools (HMDB, PHASTER, Heatmapper, CFM-ID, METAGENassist, and others).